Difference between revisions of "GARDEL:presentation"
From WikiMEG
(→Acknowledgements) |
(remove linux part) |
||
| Line 10: | Line 10: | ||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| − | | [[Image:Windows_logo.png|225px|link=http://meg.univ-amu.fr/GARDEL/download_win64.html]] || [[Image:OSX_logo.png|225px|link=http://meg.univ-amu.fr/GARDEL/download_macx | + | | [[Image:Windows_logo.png|225px|link=http://meg.univ-amu.fr/GARDEL/download_win64.html]] || [[Image:OSX_logo.png|225px|link=http://meg.univ-amu.fr/GARDEL/download_macx.html]] |
|} | |} | ||
Latest revision as of 15:22, 27 March 2023
Contents
Introduction
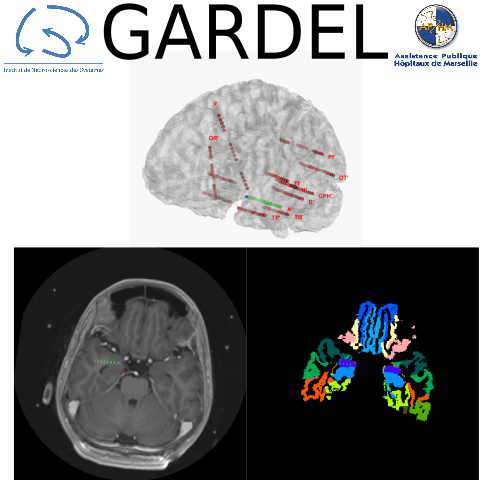
GARDEL is a tool for automatic segmentation and labelization of SEEG contacts.
Installations instructions
You can download the standalone version below
Download
 |

|
Example Data
This dataset contains data to run GARDEL or the 3Dviewer.
==>DOWNLOAD Example dataset<==
Comments/Issues
Please leave your comments/issues at samuel.medinavillalon@gmail.com
How to cite
Please cite : EpiTools, A software suite for presurgical brain mapping in epilepsy: Intracerebral EEG, Journal of Neuroscience Methods, 2018
Acknowledgements
Thanks to all Dynamap team
part of this works was financed by ANR Vibrations
Contacts
samuel.medinavillalon@gmail.com